MAKE A MEME
View Large Image
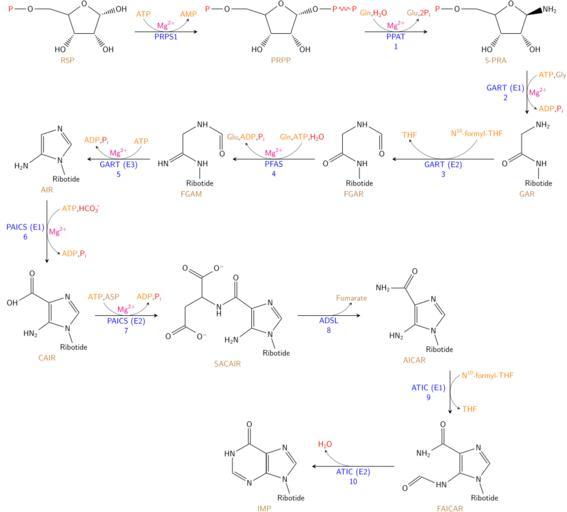
| View Original: | Nucleotides syn1.svg (849x766) | |||
| Download: | Original | Medium | Small | Thumb |
| Courtesy of: | commons.wikimedia.org | More Like This | ||
| Keywords: Nucleotides syn1.svg en The synthesis of IMP The color scheme is as follows <span style font-weight bold; ><span style color blue; >enzymes</span> <span style color orange >coenzymes</span> <span style color brown; >substrate names</span> <span style color magenta >metal ions</span> <span style color red; >inorganic molecules</span> </span> <br/>Image created using LaTeX and the generated PDF converted to SVG using http //www cityinthesky co uk/opensource/pdf2svg/ this own Krishnavedala 2014-08-06 validSVG LaTeX source code 250px <syntaxhighlight lang latex > \documentclass12pt border 1pt standalone \usepackageT1 fontenc \usepackageutf8 inputenc \usepackage chemfig chemmacros \usepackage lmodern \usetikzlibrary decorations pathmorphing \chemsetupchemformula font-shape sf format \sffamily \renewcommand \familydefault \sfdefault \renewcommand \printatom1 \ensuremath \mathsf 1 \setatomsep 2 5em \setdoublesep 6ex \setarrowdefault 2 thick \setbondstyle thick \makeatletter From http //tex stackexchange com/a/125761 Initial arguments 1 2 Same as for -U> above arrow 3 Additional label at midpoint also above arrow 4 5 6 Like 1 2 and 3 but below arrow \definearrow9 -X> \CF arrow shift nodes 7 \expandafter\draw\expandafter\CF arrow current style -CF full \CF arrow start node -- \CF arrow end node nodemidway Xarrow arctangent ; \edef\CF tmp str \ifx\ empty 1\ emptydraw none\fi \expandafter\draw\CF tmp str Xarrow arctangent arcradius \CF compound sep \CF current arrow length \ifx\ empty 8\ empty0 333\else 8\fi start angle \CF arrow current angle-90 delta angle -\ifx\ empty 9\ empty60\else 9\finode Xarrow1 start ; \edef\CF tmp str \ifx\ empty 2\ empty draw none \fi-CF full \expandafter\draw\CF tmp str Xarrow arctangent arcradius \CF compound sep \CF current arrow length \ifx\ empty 8\ empty0 333\else 8\fi start angle \CF arrow current angle-90 delta angle \ifx\ empty 9\ empty60\else 9\finode Xarrow1 end ; \edef\CF tmp str \ifx\ empty 4\ emptydraw none\fi \expandafter\draw\CF tmp str Xarrow arctangent arcradius \CF compound sep \CF current arrow length \ifx\ empty 8\ empty0 333\else 8\fi start angle \CF arrow current angle+90 delta angle \ifx\ empty 9\ empty60\else 9\finode Xarrow2 start ; \edef\CF tmp str \ifx\ empty 5\ empty draw none \fi-CF full \expandafter\draw\CF tmp str Xarrow arctangent arcradius \CF compound sep \CF current arrow length \ifx\ empty 8\ empty0 333\else 8\fi start angle \CF arrow current angle+90 delta angle -\ifx\ empty 9\ empty60\else 9\finode Xarrow2 end ; \edef\CF tmp str \if\string-\expandafter\ car\detokenize 7 \ nil-\else+\fi \CF arrow display label 1 0 \CF tmp str Xarrow1 start 2 1 \CF tmp str Xarrow1 end \CF arrow display label 3 0 5 \CF tmp str\CF arrow start node \CF arrow end node \edef\CF tmp str \if\string-\expandafter\ car\detokenize 7 \ nil+\else-\fi \CF arrow display label 4 0 \CF tmp str Xarrow2 start 5 1 \CF tmp str Xarrow2 end \CF arrow display label 6 0 5 \CF tmp str\CF arrow start node \CF arrow end node \makeatother \begin document \definesubmol\nobond - 0 2 draw none \definesubmol rb Ribotide \schemedebug true \schemestartwest \chemname \chemfig \color red P -O-- -18 5 - < HO - < OH - < OH -O- \color brown R5P \arrow -X> \color orange ATP \color orange AMP \color magenta \ch Mg 2+ \color blue PRPS1 \chemname \chemfig \color red P -O-- -18 5 - < HO - < OH - < O-0 \color red P -0 red decorate decoration snake segment length 6pt \color red P -O- \color brown PRPP \arrow -X> \color orange Gln \color red \ch H2O \color brown Glu \color red 2\ch P_i \color magenta \ch Mg 2+ \parbox 5em \centering\color blue PPAT\\1 \chemname \chemfig \color red P -O-- -18 5 - < HO - < OH - <NH_2 -O- \color brown 5-PRA \arrow -45-- -X> 0 \color orange ATP \color brown Gly 0 \color orange ADP \color red \ch P_i 0 \color magenta \ch Mg 2+ 0\parbox 5em \centering\color blue GART E1 \\2 -90 1 5 \chemname \chemfig O 30 -2- -60NH_2 - -30NH-6 1 rb \color brown GAR \arrow -X>\parbox 5em \centering\color blue GART E2 \\3 \parbox 2 75cm \color orange \ch N 10 -formyl-THF \color orange THF -2em 180 3 \chemname \chemfig O 30 6 -NH -6 rb - draw noneO -NH-- \color brown FGAR \arrow -X>\parbox 5em \centering\color blue PFAS\\4 \color orange Gln \color orange ATP \color red \ch H2O \color brown Glu \color orange ADP \color red \ch P_i \color magenta \ch Mg 2+ -2em 180 2 5 \chemname \chemfig HN 30 6 -NH -6 rb - draw noneO -NH-- \color brown FGAM \arrow -X>\parbox 5em \centering\color blue GART E3 \\5 \color orange ATP \color orange ADP \color red \ch P_i \color magenta \ch Mg 2+ -2em 180 \chemname \chemfig 5 -H_2N -N - rb - N- \color brown AIR \arrow -X> 0 \color orange ATP \color red \ch HCO3 - 0 \color orange ADP \color red \ch P_i 0 \color magenta \ch Mg 2+ 0\parbox 5em \centering\color blue PAICS E1 \\6 -90 \chemname \chemfig 5 -HN_2 -N - rb - N- - 150 -60O - 60OH \color brown CAIR \arrow -X> \color orange ATP \color brown ASP \color orange ADP \color red \ch P_i \color magenta \ch Mg 2+ \parbox 5em \centering\color blue PAICS E2 \\7 \chemname \chemfig O 30 6 -O - - draw none- draw none\chembelow N H - 30 2O - -36 5 -H_2N -N - rb - N- - - 60O - -60O - -- \color brown SACAIR \arrow -X> \color brown Fumarate \parbox 5em \centering\color blue ADSL\\8 \quad \chemname \chemfig 5 -HN_2 -N - rb - N- - 120 -60O - 60NH_2 \color brown AICAR \arrow -50-- -X> 0\parbox 2 75cm \color orange \ch N 10 -formyl-THF 0 \color orange THF 0\parbox 5em \centering\color blue ATIC E1 \\9 -90 1 5 \chemname \chemfig 5 -HN- -60 60O -N - rb - N- - 150 -60O - 60NH_2 \color brown FAICAR \arrow -X>\parbox 5em \centering\color blue ATIC E2 \\10 \color red \ch H2O -2em 180 2 5 \chemname \chemfig 6 N- 5 -N - rb - N- -- O - 2HN- 2 \color brown IMP \schemestop \end document </syntaxhighlight> cob Genetics Nucleotides Cc-zero | ||||